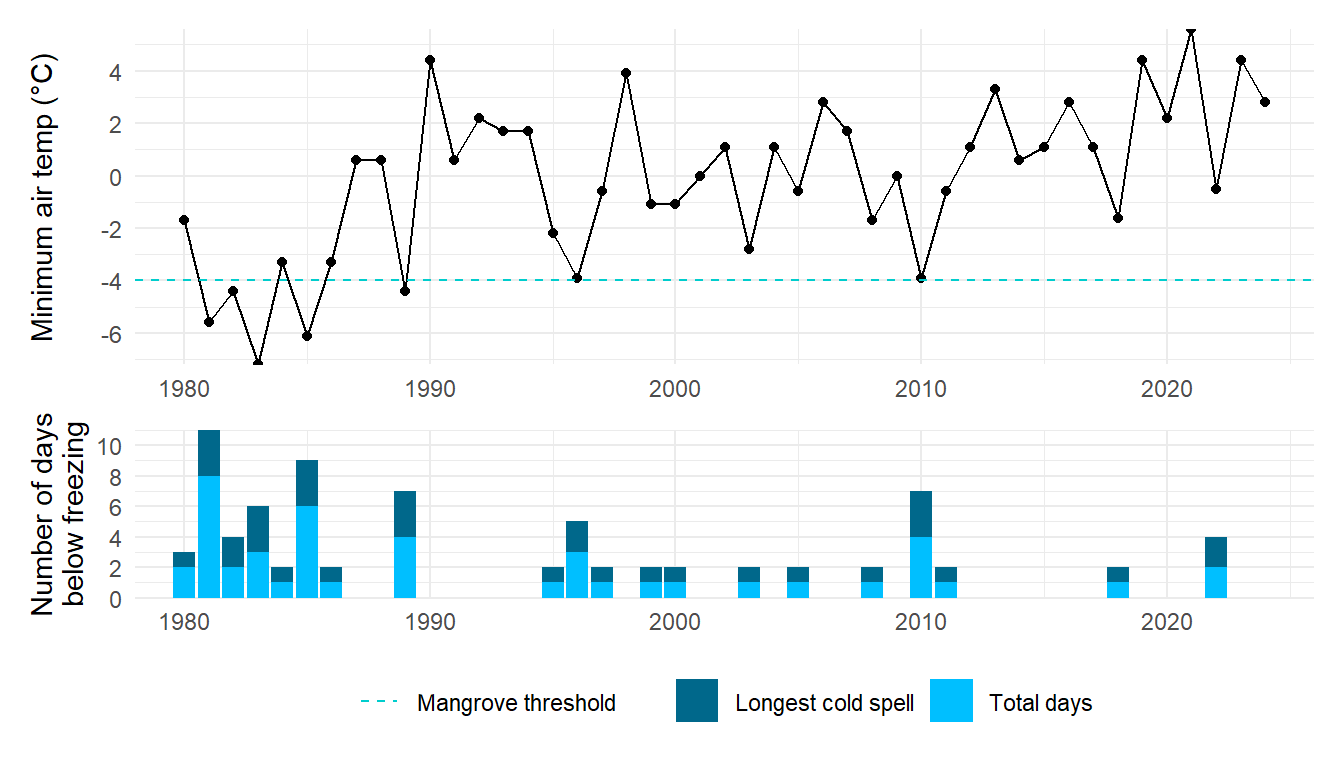
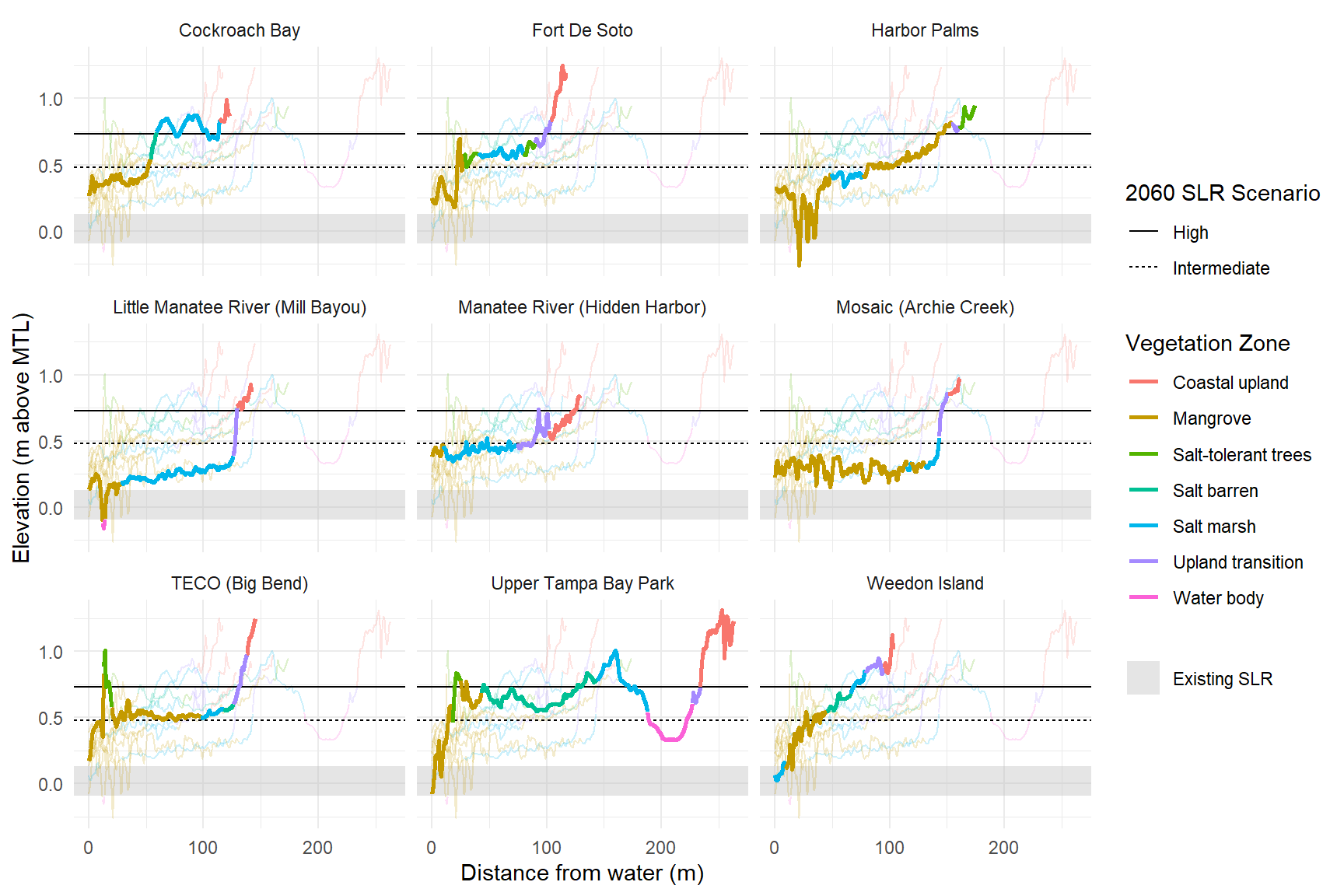
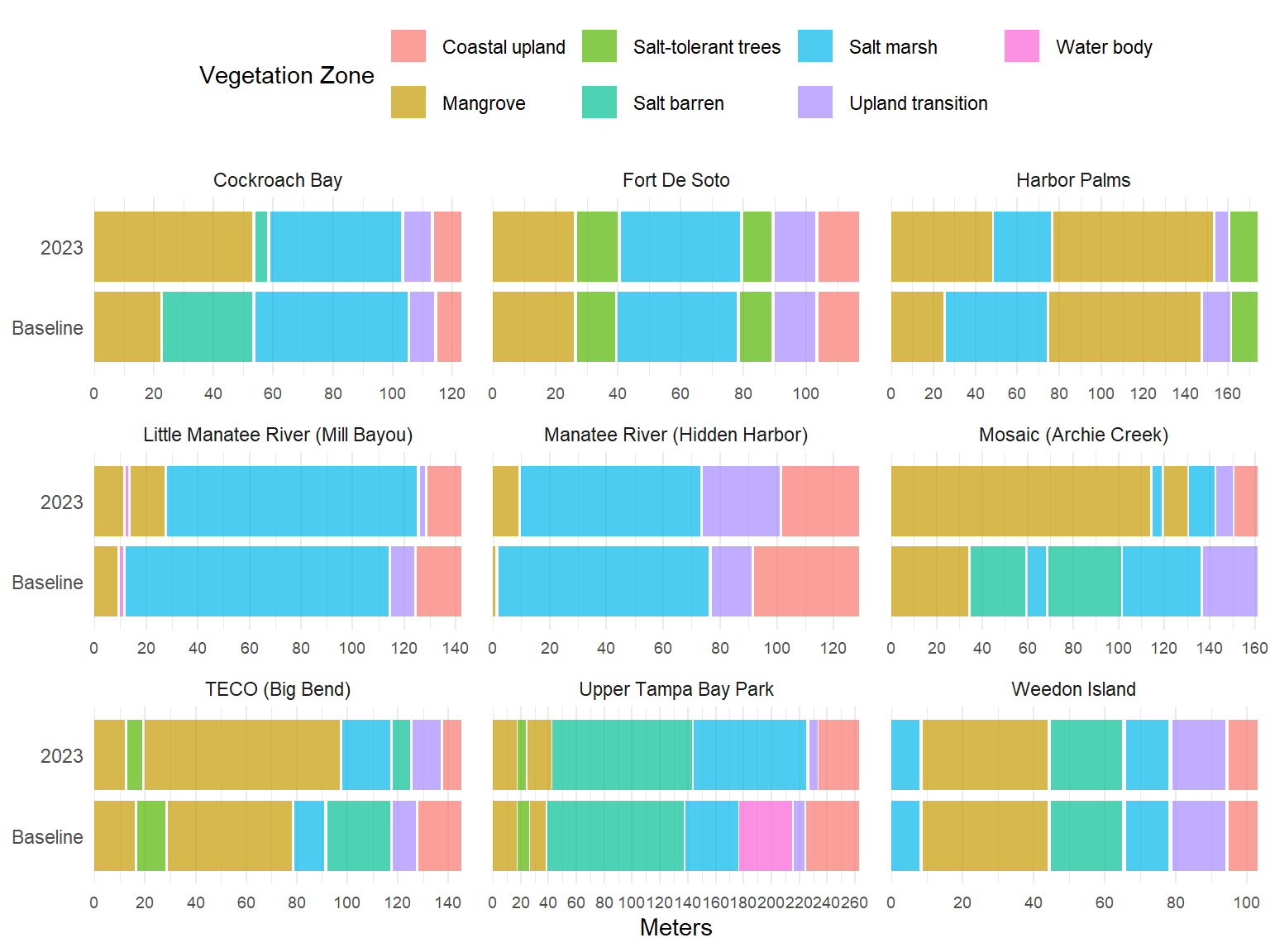
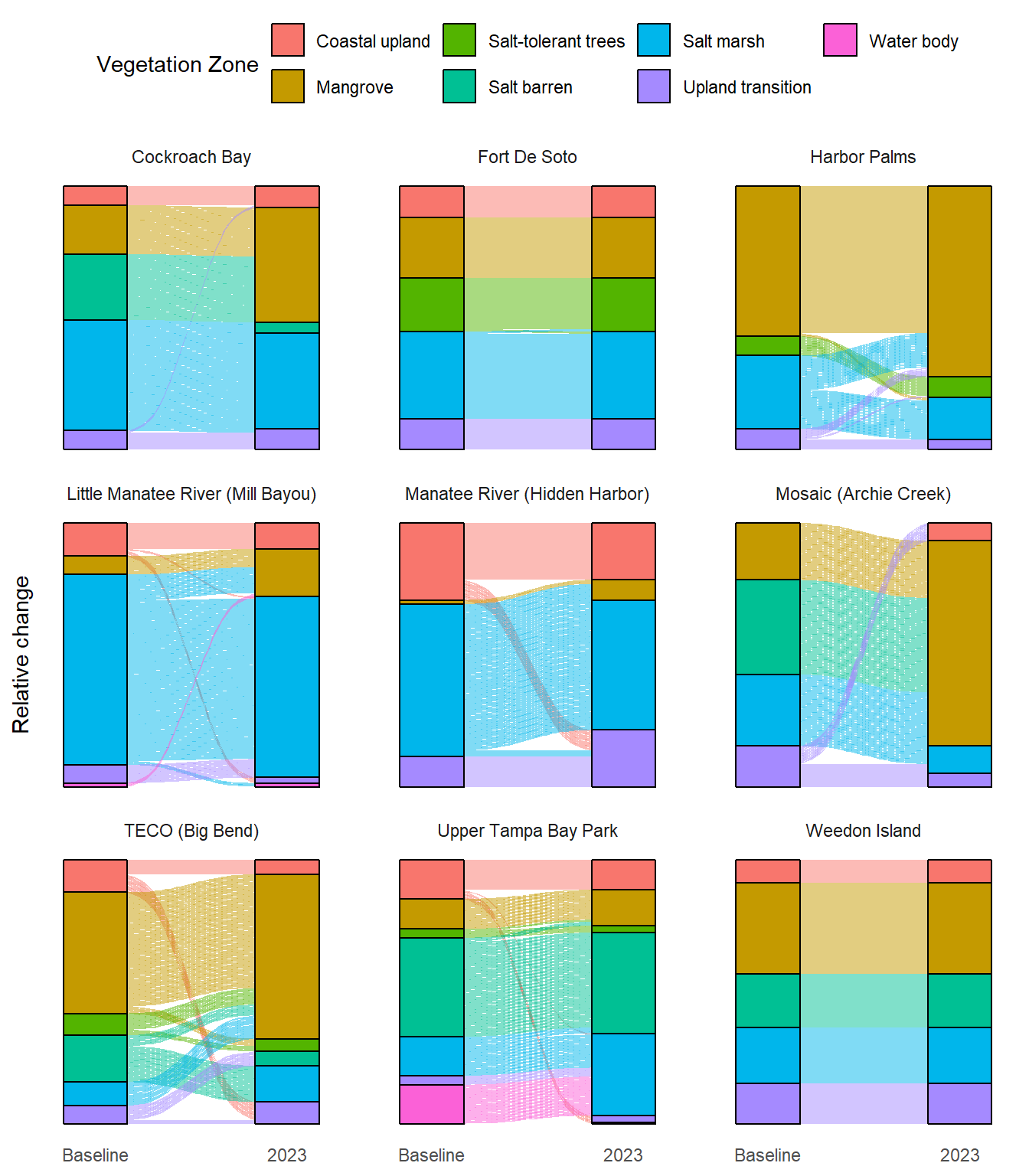
# # accretion rates, based on RTK elevation diff per year
# accrt <- vegdatele %>%
# mutate(
# yr = year(mdy(Date))
# ) %>%
# summarise(
# meanele = mean(Elevation_NAVD88, na.rm = T),
# .by = c(Site, yr)
# ) %>%
# summarise(
# elediff = diff(meanele),
# yrdiff = diff(yr),
# .by = Site
# ) %>%
# mutate(
# accrate = elediff / yrdiff
# ) %>%
# select(Site, accrate)
ele <- vegdatelezcomp %>%
filter(Sample_set == 3) %>%
select(Site, Simplified_zone_name, Meter, Elevation_MTL) %>%
distinct() |>
mutate(
zone_group = cumsum(Simplified_zone_name != lag(Simplified_zone_name, default = first(Simplified_zone_name))) + 1,
.by = Site
)
interpolate_transect <- function(df) {
# Process each site separately
df_interpolated <- df %>%
group_by(Site) %>%
do({
site_data <- .
# Create the new meter sequence at 0.25m intervals
meter_seq <- seq(from = min(site_data$Meter),
to = max(site_data$Meter),
by = 0.1)
# Create a new dataframe with the interpolated meter values
new_df <- data.frame(
Site = unique(site_data$Site),
Meter = meter_seq
)
# Interpolate elevation using linear interpolation
new_df$Elevation_MTL <- approx(x = site_data$Meter,
y = site_data$Elevation_MTL,
xout = meter_seq,
method = "linear")$y
# For categorical variables, use "last observation carried forward"
# This assumes zones are continuous until they change
# First, create a mapping of meter to zone values
zone_df <- site_data %>%
select(Meter, Simplified_zone_name, zone_group)
# Merge and fill forward the categorical values
new_df <- new_df %>%
left_join(zone_df, by = "Meter") %>%
arrange(Meter) %>%
fill(Simplified_zone_name, zone_group, .direction = "down")
new_df
}) %>%
ungroup()
return(df_interpolated)
}
toplo <- interpolate_transect(ele)
toplo2 <- toplo |>
unite(zone_group, c('Site', 'zone_group'), sep = '_')
lns <- tibble(
Scenario = c('High', 'Intermediate'),
elev = c(0.73, 0.48)
)
rects <- tibble(
Site = unique(toplo$Site),
ymin = -0.095,
ymax = 0.129
)
p <- ggplot(toplo, aes(x = Meter, y = Elevation_MTL, color = Simplified_zone_name, group = zone_group)) +
geom_rect(data = rects, aes(ymin = ymin, ymax = ymax, fill = 'Existing SLR', xmin = -Inf, xmax = Inf), alpha = 0.4, inherit.aes = F) +
geom_line(data = toplo2, aes(x = Meter, y = Elevation_MTL, group =
zone_group, color = Simplified_zone_name), alpha = 0.2) +
geom_hline(data = lns, aes(yintercept = elev, linetype = Scenario), color = 'black') +
geom_line(size = 1) +
scale_fill_manual(values = 'grey') +
facet_wrap(~ Site) +
theme_minimal() +
# theme(
# legend.position = "bottom"
# ) +
labs(
x = "Distance from water (m)",
y = "Elevation (m above MTL)",
color = "Vegetation Zone",
fill = NULL,
linetype = '2060 SLR Scenario'
)
p