Code
library (tidyverse)library (randomForest)library (janitor)library (caret)library (zoo)library (here)load (file = here ('data/vegdat.RData' ))# colors <- function (n) {= seq (15 , 375 , length = n + 1 )hcl (h = hues, l = 65 , c = 100 )[1 : n]<- vegdat %>% select (zone_name_simp) %>% distinct () %>% arrange (zone_name_simp) %>% na.omit () %>% pull ()<- cols (length (levs))names (colin) <- levs# smoothing function for prediction post-processing <- function (prds, win = 3 , align = 'left' , fillna = F){<- levels (prds)<- as.character (prds)<- rollapply (width = win, FUN = function (x){names (sort (table (x), decreasing = TRUE )[1 ])align = align, fill = NA )if (fillna)<- ifelse (is.na (out), prds, out)<- factor (out, levels = levs)return (out)# species in all three samples <- vegdat %>% select (sample, species) %>% distinct () %>% mutate (present = 1 %>% pivot_wider (names_from = sample, values_from = present, values_fill = 0 ) %>% mutate (allpresent = ifelse (` 1 ` + ` 2 ` + ` 3 ` == 3 , 1 , 0 )%>% filter (allpresent == 1 ) %>% pull (species) %>% sort ()
This analysis is a proof of concept for predicting vegetation zones in phase 2 sampling, where complete vegetation plots were sampled but no zone or elevation information was recorded. The predictions use a random forest model trained on the phase 1 data and tested on phase 3 data. The model used species basal percent cover, meter mark, and site as predictors. Only species shared across all three phases were included. The model predictions compared to actual zones for phase 1 (training) and phase 3 (validation) are shown for comparison, including model confusion matrices and accuracy statistics. Model predictions using smoothed results are also evaluated, where meter marks are assigned the majority zone within a rolling window of width 5.
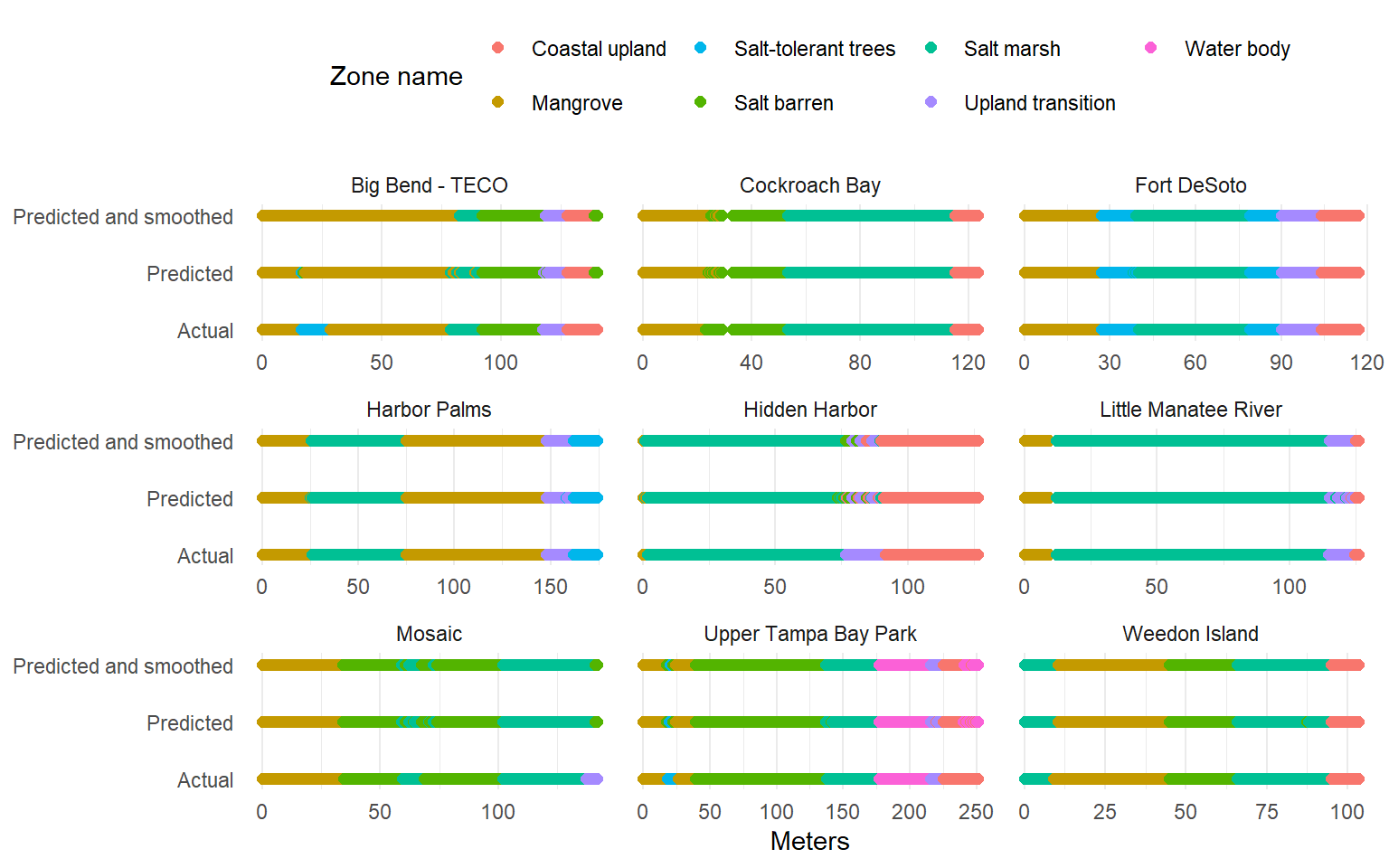
Phase 1 training data
Code
# prep for rf, use first phase as training <- vegdat %>% filter (species %in% shrspp) %>% filter (sample == 1 ) %>% select (%>% summarise (pcent_basal_cover = mean (pcent_basal_cover, na.rm = T),.by = c (site, zone_name_simp, meter, species)%>% filter (! is.na (pcent_basal_cover)) %>% pivot_wider (names_from = species, values_from = pcent_basal_cover, values_fill = 0 ) %>% clean_names () %>% mutate_if (is.character, as.factor)# rf mod <- randomForest (zone_name_simp ~ ., data = trndat, importance = TRUE , ntree = 500 , keep.inbag = T)# data for checking results <- trndat %>% mutate (zone_name_simpprd = predict (rfmod, .)%>% select (%>% mutate (zone_name_simpprdsmth = smooth_fun (zone_name_simpprd, align = 'center' , win = 5 , fillna = T),.by = site
Code
# visualize training results <- trndatchk %>% pivot_longer (cols = - c (site, meter),names_to = 'typ' ,values_to = 'zone_name_simp' %>% filter (! is.na (zone_name_simp)) %>% mutate (typ = case_when (grepl ('prd$' , typ) ~ 'Predicted' , grepl ('prdsmth$' , typ) ~ 'Predicted and smoothed' , ~ 'Actual' ggplot (toplo, aes (x = meter, y = typ, color = zone_name_simp)) + geom_point (size = 2 ) + facet_wrap (~ site, scales = 'free_x' ) + scale_color_manual (values = colin) + scale_y_discrete (expand = c (0.1 , 0 )) + theme_minimal () + theme (legend.position = 'top' ,panel.grid.major.y = element_blank ()+ labs (x = 'Meters' , y = NULL , color = 'Zone name'
Confusion matrix and accuracy statistic for model predictions compared to actual zones (reference).
Code
<- confusionMatrix (trndatchk$ zone_name_simpprd, trndatchk$ zone_name_simp)$ table
Reference
Prediction Coastal upland Mangrove Salt-tolerant trees Salt barren
Coastal upland 195 0 0 0
Mangrove 0 646 26 7
Salt-tolerant trees 0 0 76 0
Salt barren 3 2 10 445
Salt marsh 0 5 2 6
Upland transition 1 0 0 0
Water body 17 0 0 0
Reference
Prediction Salt marsh Upland transition Water body
Coastal upland 0 9 0
Mangrove 7 0 0
Salt-tolerant trees 0 1 0
Salt barren 12 10 0
Salt marsh 904 15 0
Upland transition 0 119 0
Water body 0 0 78
Code
Confusion matrix and accuracy statistic for smoothed model predictions compared to actual zones (reference).
Code
<- confusionMatrix (trndatchk$ zone_name_simpprdsmth, trndatchk$ zone_name_simp)$ table
Reference
Prediction Coastal upland Mangrove Salt-tolerant trees Salt barren
Coastal upland 191 0 0 0
Mangrove 0 645 29 7
Salt-tolerant trees 0 0 75 0
Salt barren 3 2 8 446
Salt marsh 0 6 2 5
Upland transition 1 0 0 0
Water body 21 0 0 0
Reference
Prediction Salt marsh Upland transition Water body
Coastal upland 0 7 0
Mangrove 7 0 0
Salt-tolerant trees 0 0 0
Salt barren 5 12 0
Salt marsh 911 10 0
Upland transition 0 125 0
Water body 0 0 78
Code
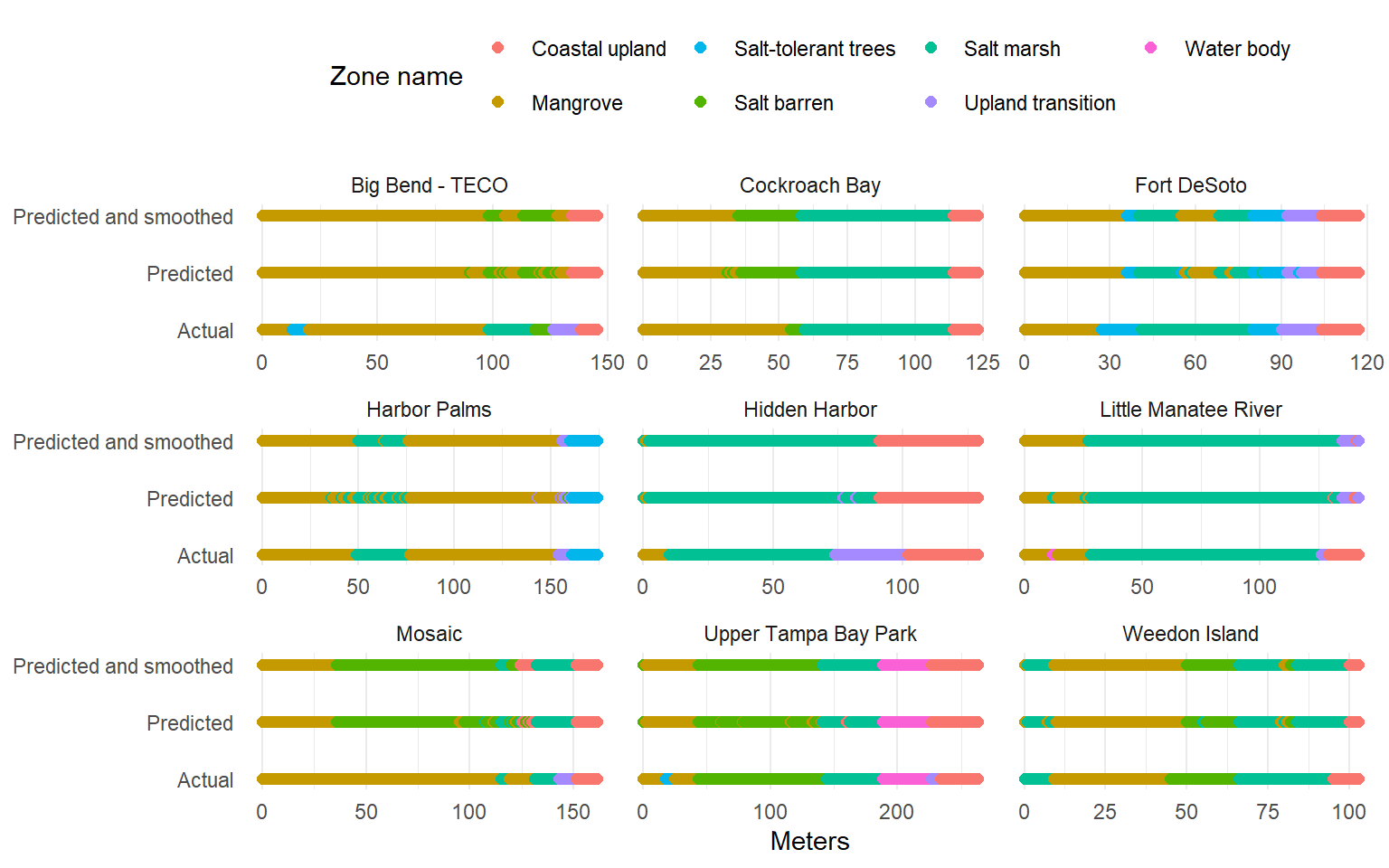
Phase 3 testing data
Code
# test dataset on phase 3 <- vegdat %>% filter (sample == 3 ) %>% select (%>% summarise (pcent_basal_cover = mean (pcent_basal_cover, na.rm = T),.by = c (site, zone_name_simp, meter, species)%>% pivot_wider (names_from = species, values_from = pcent_basal_cover, values_fill = 0 ) %>% clean_names () %>% mutate_if (is.character, as.factor)<- tstdat %>% mutate (zone_name_simpprd = predict (rfmod, .)%>% select (%>% mutate (zone_name_simpprdsmth = smooth_fun (zone_name_simpprd, align = 'center' , win = 5 , fillna = T),.by = site
Code
# visualize testing results <- tstdatchk %>% pivot_longer (cols = - c (site, meter),names_to = 'typ' ,values_to = 'zone_name_simp' %>% filter (! is.na (zone_name_simp)) %>% mutate (typ = case_when (grepl ('prd$' , typ) ~ 'Predicted' , grepl ('prdsmth$' , typ) ~ 'Predicted and smoothed' , ~ 'Actual' ggplot (toplo, aes (x = meter, y = typ, color = zone_name_simp)) + geom_point (size = 2 ) + facet_wrap (~ site, scales = 'free_x' ) + scale_color_manual (values = colin) + scale_y_discrete (expand = c (0.1 , 0 )) + theme_minimal () + theme (legend.position = 'top' ,panel.grid.major.y = element_blank ()+ labs (x = 'Meters' , y = NULL , color = 'Zone name'
Confusion matrix and accuracy statistic for model predictions compared to actual zones (reference).
Code
<- confusionMatrix (tstdatchk$ zone_name_simpprd, tstdatchk$ zone_name_simp)$ table
Reference
Prediction Coastal upland Mangrove Salt-tolerant trees Salt barren
Coastal upland 108 4 0 0
Mangrove 0 407 23 13
Salt-tolerant trees 0 0 27 0
Salt barren 0 103 0 116
Salt marsh 10 18 2 5
Upland transition 6 1 0 0
Water body 0 0 0 0
Reference
Prediction Salt marsh Upland transition Water body
Coastal upland 3 22 0
Mangrove 33 8 0
Salt-tolerant trees 1 3 0
Salt barren 14 3 0
Salt marsh 352 26 2
Upland transition 0 17 0
Water body 0 0 39
Code
Confusion matrix and accuracy statistic for smoothed model predictions compared to actual zones (reference).
Code
<- confusionMatrix (tstdatchk$ zone_name_simpprdsmth, tstdatchk$ zone_name_simp)$ table
Reference
Prediction Coastal upland Mangrove Salt-tolerant trees Salt barren
Coastal upland 106 7 0 0
Mangrove 0 411 23 5
Salt-tolerant trees 0 0 28 0
Salt barren 0 104 0 125
Salt marsh 11 11 1 4
Upland transition 7 0 0 0
Water body 0 0 0 0
Reference
Prediction Salt marsh Upland transition Water body
Coastal upland 1 22 0
Mangrove 27 8 2
Salt-tolerant trees 0 3 0
Salt barren 14 2 0
Salt marsh 361 28 0
Upland transition 0 16 0
Water body 0 0 39
Code
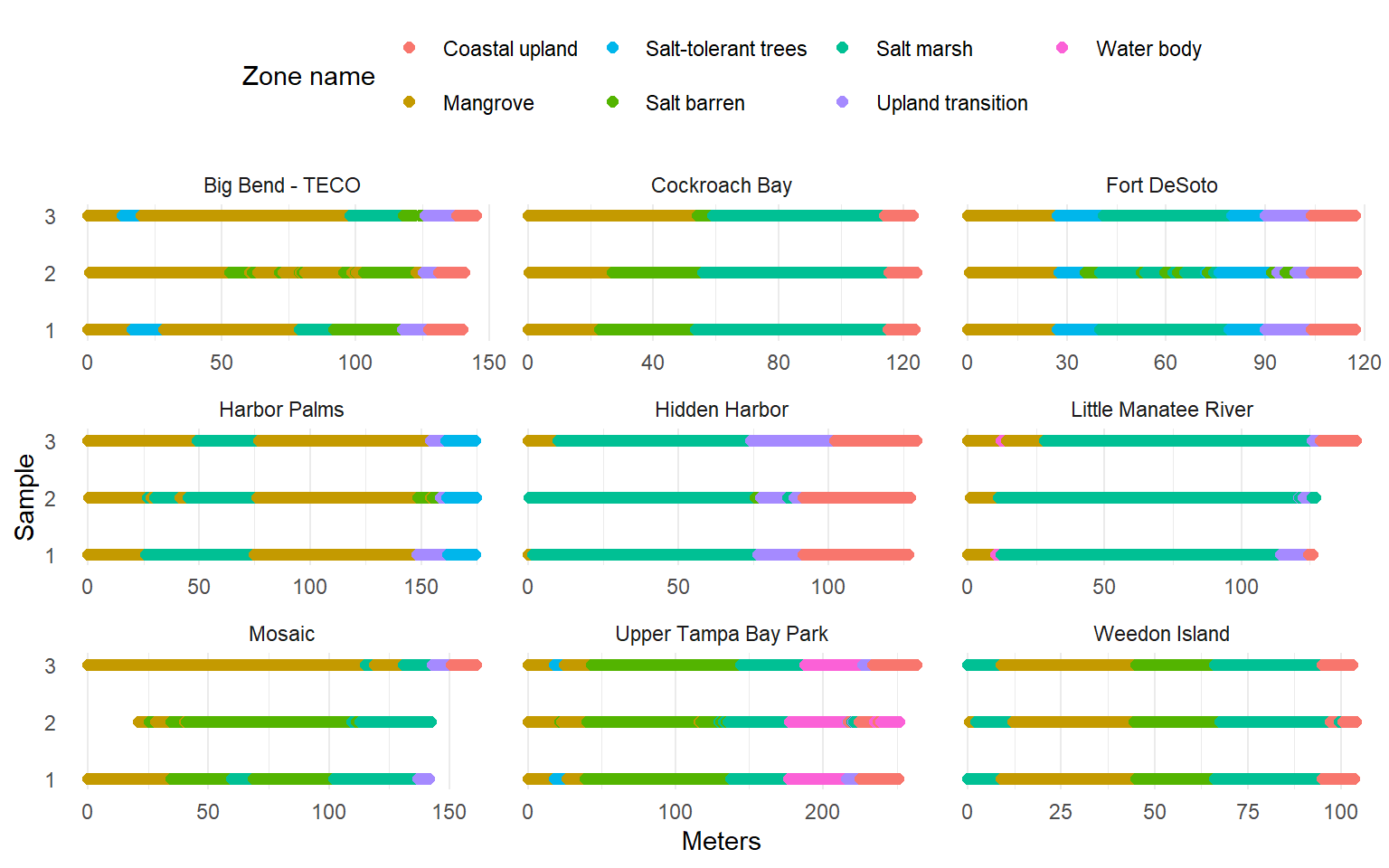
Phase 2 predictions
Code
# predict sample 2 <- vegdat %>% filter (sample == 2 ) %>% select (%>% summarise (pcent_basal_cover = mean (pcent_basal_cover, na.rm = T),.by = c (site, zone_name_simp, meter, species)%>% pivot_wider (names_from = species, values_from = pcent_basal_cover, values_fill = 0 ) %>% clean_names () %>% mutate_if (is.character, as.factor)<- toprd %>% mutate (sample = 2 ,zone_name_simp2 = predict (rfmod, newdata = .)%>% mutate (zone_name_simp2 = smooth_fun (zone_name_simp2, align = 'center' , win = 5 , fillna = T),.by = site%>% select (site, sample, meter, zone_name_simp2) %>% mutate (site = as.character (site),zone_name_simp2 = as.character (zone_name_simp2)<- vegdat %>% group_nest (site, sample, zone_name_simp, meter) %>% left_join (tomrg, by = c ('site' , 'sample' , 'meter' )) %>% mutate (zone_name_simp = case_when (is.na (zone_name_simp) ~ zone_name_simp2, ~ zone_name_simp%>% select (- zone_name_simp2) %>% unnest ('data' ) %>% arrange (sample, site, meter, species)
Code
<- vegdat2 %>% select (site, sample, zone_name_simp, meter) %>% distinct () %>% mutate (sample = factor (sample)ggplot (toplo, aes (x = meter, y = sample, color = zone_name_simp)) + geom_point (size = 2 ) + facet_wrap (~ site, scales = 'free_x' ) + scale_color_manual (values = colin) + scale_y_discrete (expand = c (0.1 , 0 )) + theme_minimal () + theme (legend.position = 'top' ,panel.grid.major.y = element_blank ()+ labs (x = 'Meters' , y = 'Sample' , color = 'Zone name'